ChIC/CUT&RUN Assay Kit Overview
Related Products:
- CUT&RUN-validated antibody: For best results with your samples
- CUT&RUN Spike-In Control: Evaluate CUT&RUN assay datasets confidently for comparison
CUT&RUN (Cleavage Under Targets & Release Using Nuclease) is an epigenetic method used to investigate the genome-wide distribution of various chromatin-associated proteins and their modifications1-3. CUT&RUN is a derivative of chromatin immunocleavage (ChIC). CUT&RUN is similar to chromatin immunoprecipitation (ChIP), in that it utilizes an antibody to target chromatin-associated marks and proteins, but requires less sample material and less sequencing depths than ChIP.
In CUT&RUN, a protein of interest is tagged with an antibody and bound to the chromatin in intact cells. Then, a micrococcal nuclease (MNase) is used to cleave the DNA specifically at the binding sites of the protein of interest. The released fragments are purified, sequenced, and mapped to the reference genome to determine the protein’s binding sites. Unlike ChIP, CUT&RUN does not require crosslinking of the protein to the DNA, which can introduce artifacts.
CUT&RUN is a valuable tool for studying chromatin-associated proteins because it is sensitive, specific, and requires fewer cells than ChIP, making it ideal for identifying binding patterns of chromatin-associated proteins such as transcription factors or histone modifications genome-wide. Chromatin-associated proteins play critical roles in regulating various cellular processes such as gene expression, DNA replication, DNA repair, and cell differentiation. Understanding the binding patterns of these proteins can provide insight into how these cellular processes are regulated.
References:
1. Schmid, M. et al. Mol Cell, 16(1): 147-157 (2004)
2. Skene, P.J. et al. (2017) Elife 6, e21856
3. Skene, P.J. et al. (2018) Nat. Protoc., 13, 1006-1019
ChIC/CUT&RUN Assay Kit Advantages:
- Compatible with 5,000 to 500,000 cells
- Complete kit with optimized protocol
- Developed for genome-wide chromatin-associated protein profiling
How the ChIC/CUT&RUN Assay Works

CUT&RUN vs. CUT&Tag vs. ChIP-Seq
| CUT&RUN | CUT&Tag | ChIP-Seq | |
|---|---|---|---|
| Performed Under Native Conditions? | Yes | Yes | No |
| Chromatin Fragmentation Method | MNase digestion | Tn5-based tagmentation | Sonication |
| Cell Number Requirements | 500,000 cells | 5,000-500,000 cells | 1-10 million cells |
| Sequencing Depth Required * | 8 million reads † | 2 million reads ** | 20-50 million reads |
| Integrated Library Preparation? | No, separate library prep required | Yes, uses tagmentation | No, separate library prep required |
| Compatible Targets | Wide range of histone modifications, transcription factors, and co-factors | Primarily histone modifications, some transcription factors and co-factors | Wide range of histone modifications, transcription factors, and co-factors |
| Workflow Length | 1-2 days | 1-2 days | 2-3 days |
* Kaya-Okur et al. Nature Communications (2019) 10:1930
** For less abundant targets of interest, 8 to 10 million reads are recommended
† 25 million reads are recommended for transcription factor targets
ChIC/CUT&RUN Assay Kit Contents
The ChIC/CUT&RUN Assay Kit components are shipped at two temperatures, with one box on dry ice for components to be stored at -20°C, and a second box at room temperature for components that are not to be frozen and must be stored at 4°C. Please store components according to the storage conditions in the manual after first use. All reagents are guaranteed stable for 6 months from date of receipt when stored properly.
- Protease Inhibitor Cocktail, 500 µl, store at -20°C
- ChIC/CUT&RUN pAG-MNase, store at -20°C
- Glycogen 20 mg/ml, store at -20°C
- Histone H3K4me3 pAb, 10 µg, 1 mg/mL store at -20°C
- Negative Control IgG, Rabbit, store at -20°C
- 1 M Spermidine, store at -20°C
- Nuclei Isolation Buffer, store at 4°C
- Concanavalin A Beads, store at 4°C
- 0.1 M CaCl2, store at 4°C
- Stop Solution, store at 4°C
- 1X Binding Buffer, store at 4°C
- Dig-Wash Buffer, store at 4°C
- 0.5 M EDTA, store at Room temperature
- DNA Purification Wash Buffer, store at Room temperature
- DNA Purification Elution Buffer, store at Room temperature
- DNA Purification Binding Buffer, store at Room temperature
- 3M Sodium Acetate, store at Room temperature
- DNA Purification Columns SF, store at Room temperature
- 0.2 mL Strip Tubes, Attached Caps, store at Room temperature
- RNase A, store at -20°C
- 5% Digitonin, store at -20°C
ChIC/CUT&RUN Assay Kit Data
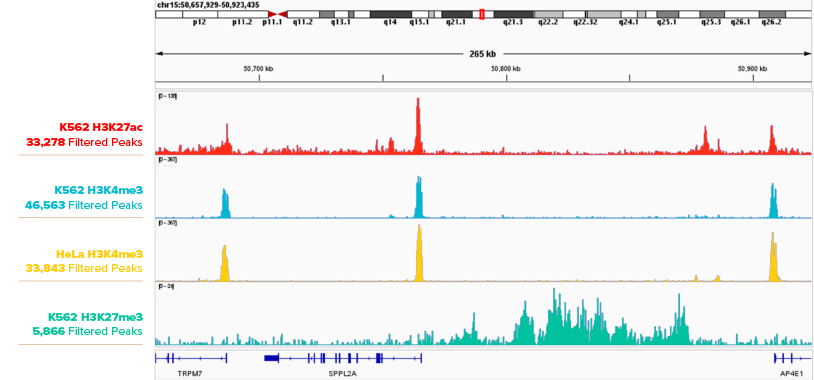
Figure 1. CUT&RUN Profiling Histone Marks in K562 Cells.
IGB browser tracks are shown for 500,000 K562 cell CUT&RUN peaks with Active Motif antibodies H3K27ac (Cat. No. 91193), H3K4me3 (Cat. No. 91263) and H3K27me3 (Cat. No. 39155).
Figure 2. ETS1 CUT&RUN Peak Profiles Are Similar to ETS1 ChIP-Seq Profiles.
IGB browser tracks for ChIP-Seq (top two tracks) in DIPTG cells and CUT&RUN (bottom two tracks) in K562 cells assays targeting ETS1 (Cat. No. 39580). Sequence logos from Homer analysis (to the right of the tracks) show motif enrichment for the ETS1 motif in both sample sets.
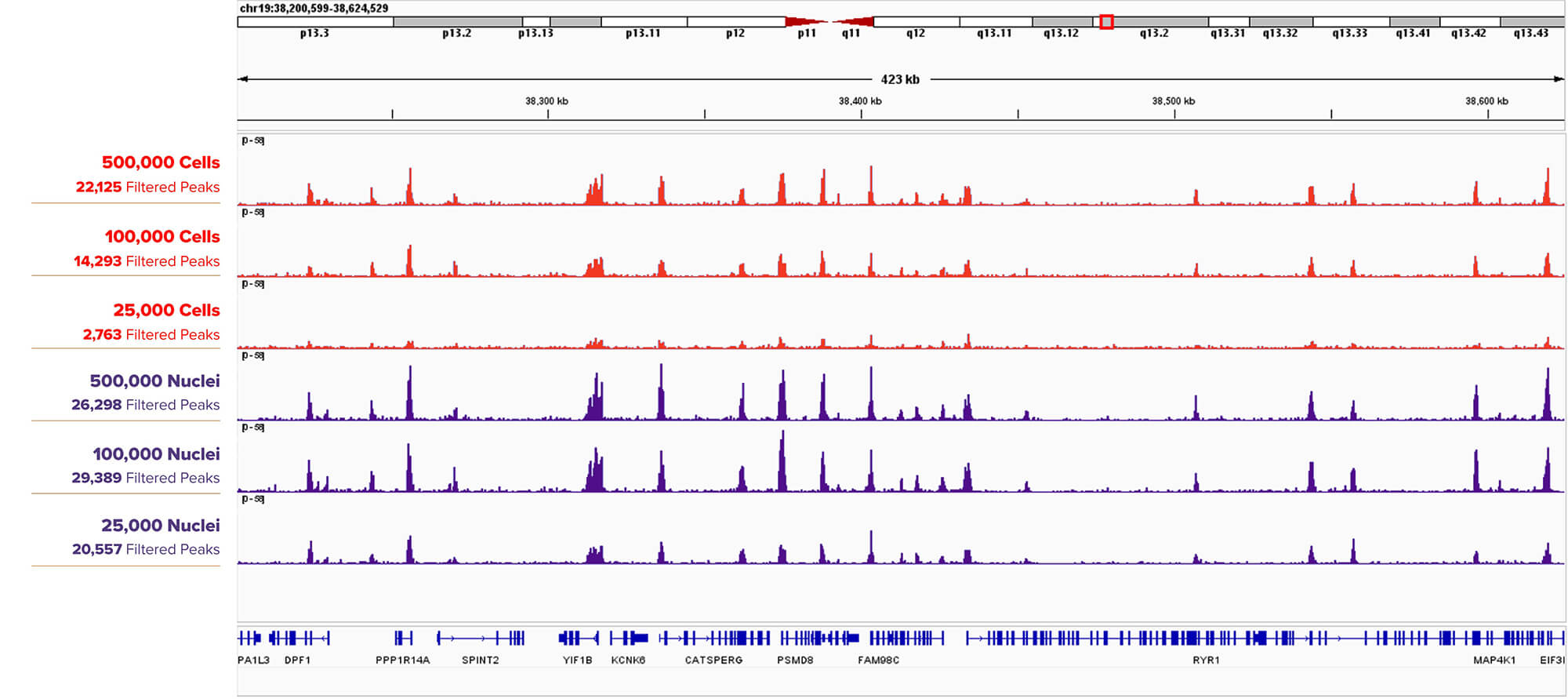
Figure 3. CUT&RUN H3K4me3 Assay Results Comparing Titrations of K562 Cells versus Nuclei.
500,000, 100,000, 25,000 K562 Cells (top three tracks) or Nuclei (bottom three tracks) were assayed in CUT&RUN with Active Motif’s H3K4me3 antibody (Cat. No. 39060). Filtered Peaks remain higher at lower input for nuclei than cells, and the browser tracks for nuclei show stronger peaks for each input amount when compared to cells.
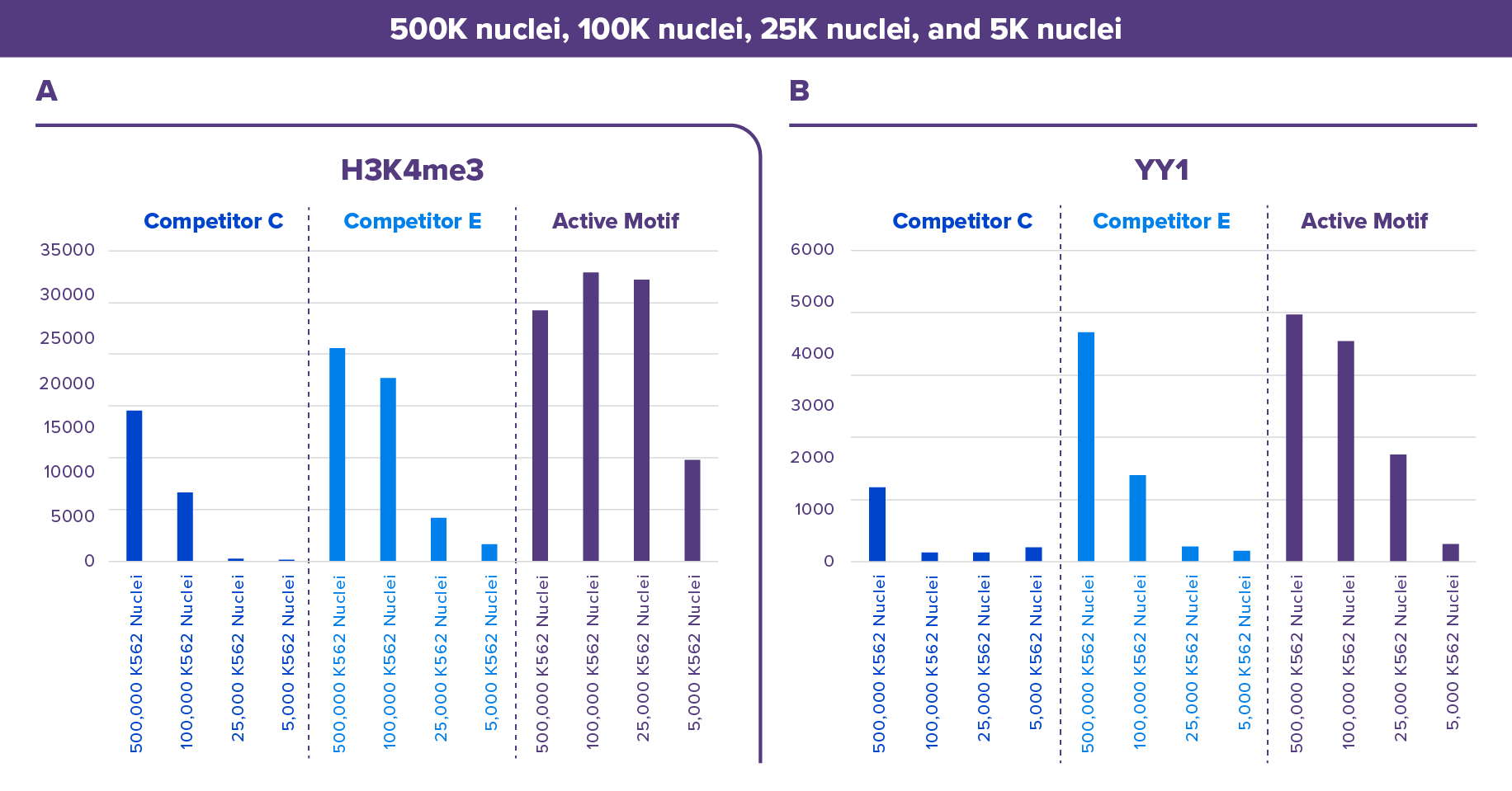
Figure 4. Active Motif’s CUT&RUN Assay Kit Outperforms the Competition.
Peak scores are plotted in the bar graphs for 500,000, 100,000, 25,000, and 5,000 K562 nuclei assayed in CUT&RUN Kits from either Active Motif, Competitor C, and Competitor E for YY1 (Active Motif Cat. No. 61980) and H3K4me3 (Active Motif Cat. No. 39916).
Figure 5. Active Motif’s CUT&RUN Assay Kit is Compatible with 5,000 Nuclei for Histone Marks.
5,000 fresh K562 nuclei were assayed using Active Motif’s CUT&RUN Assay Kit or Competitor E or Competitor C CUT&RUN Assay Kits using Active Motif's H3K4me3 antibody (Cat. No. 91263). Active Motif’s CUT&RUN Assay Kit yielded more robust results than the competition.
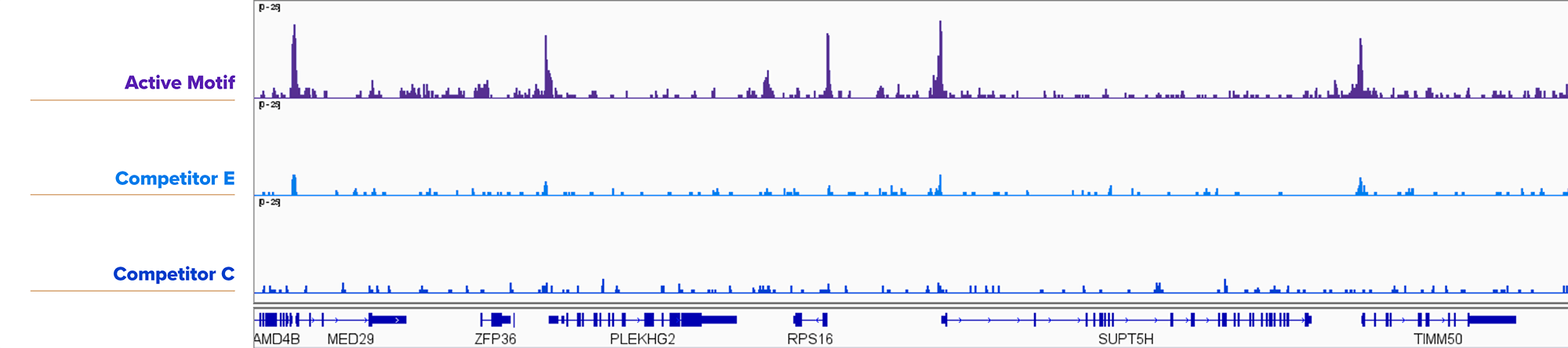
Figure 6. Active Motif’s CUT&RUN Assay Kit is Compatible with 25,000 Nuclei for Transcription Factors.
25,000 fresh K562 nuclei were assayed using Active Motif’s CUT&RUN Assay Kit or Competitor E or Competitor C CUT&RUN Assay Kits using Active Motif's YY1 antibody (Cat. No. 61980). Active Motif’s CUT&RUN Assay Kit yielded more robust results than the competition.
ChIC/CUT&RUN Assay Kit FAQs
The nuclei pellet is not visible, is it there?
The pellet may not be visible. Be sure to position tubes with caps oriented in centrifuge so that you know where the pellet should be in the tube, and proceed carefully with the next steps.
What is the recommended sequencing depth?
25 million reads for transcription factors. For some histone marks you may go down to 2 million reads.
What is the recommended read length for CUT&RUN?
The recommended read length for CUT&RUN is PE38.
How many cells should I use for transcription factor targets?
We recommend 500,000 cells and a CUT&RUN validated antibody.
Why is there high background in the data?
Be sure to start with viable cells. Check that the cells/nuclei are intact by an automated cell counting method such as the Countess II or by Trypan Blue staining and a hemocytometer.
Will the negative control produce a library?
Yes. The negative control IgG will produce a library.
SPRI beads are not pelleting towards the bottom or are taking too long to pellet, what can I do?
Using a magnet, start by pelleting the SPRI beads towards the top of the tube, and gradually work your way down to the bottom of the tubes. Then, place the tubes on a magnetic plate.
Which Active Motif antibodies have been published for CUT&RUN?
The antibodies listed here have been used for CUT&RUN in publications. These have not been validated for CUT&RUN by Active Motif.
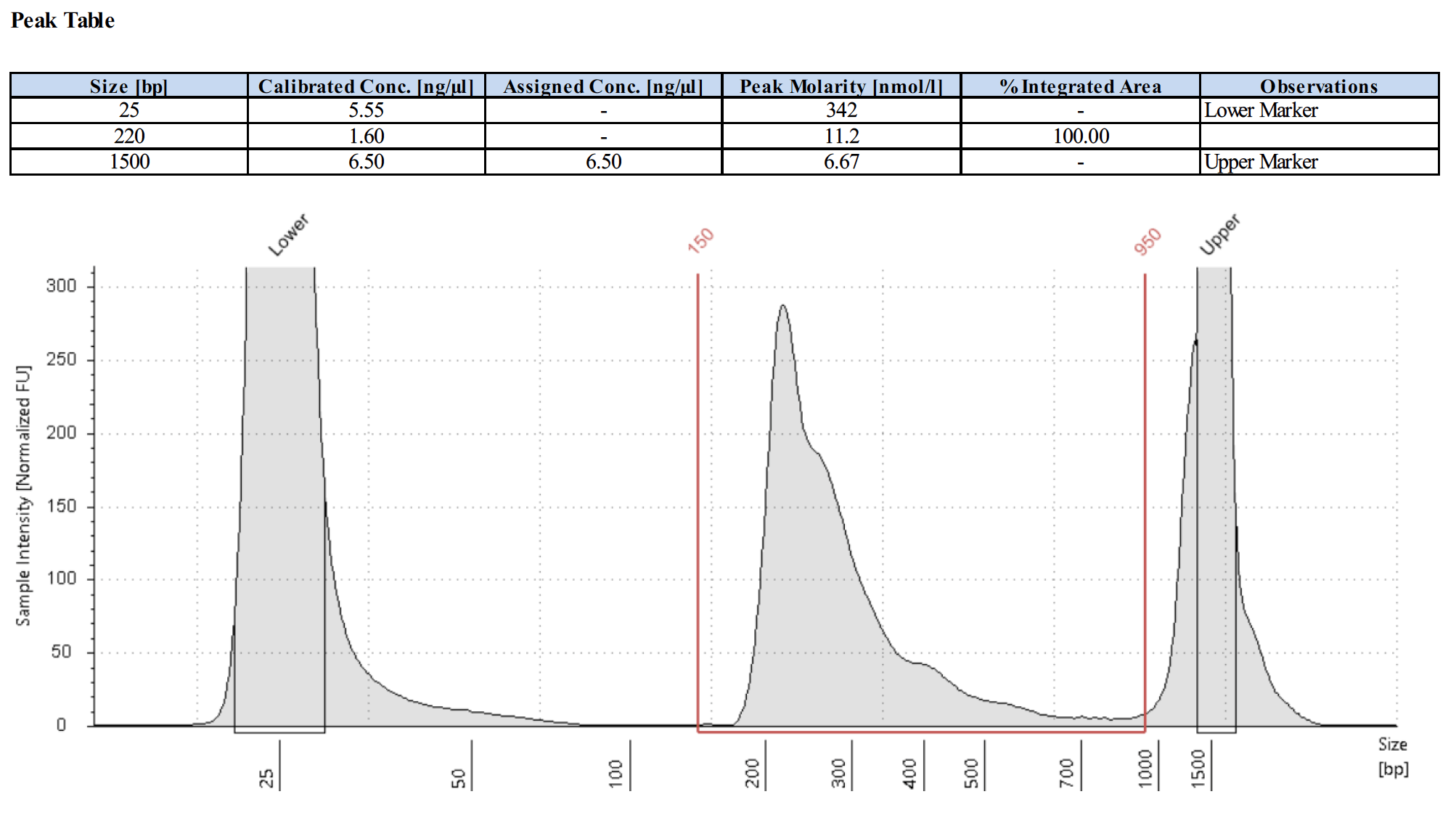
What do library traces look like for the positive control antibody?
Here are 4 examples of the positive control library traces.




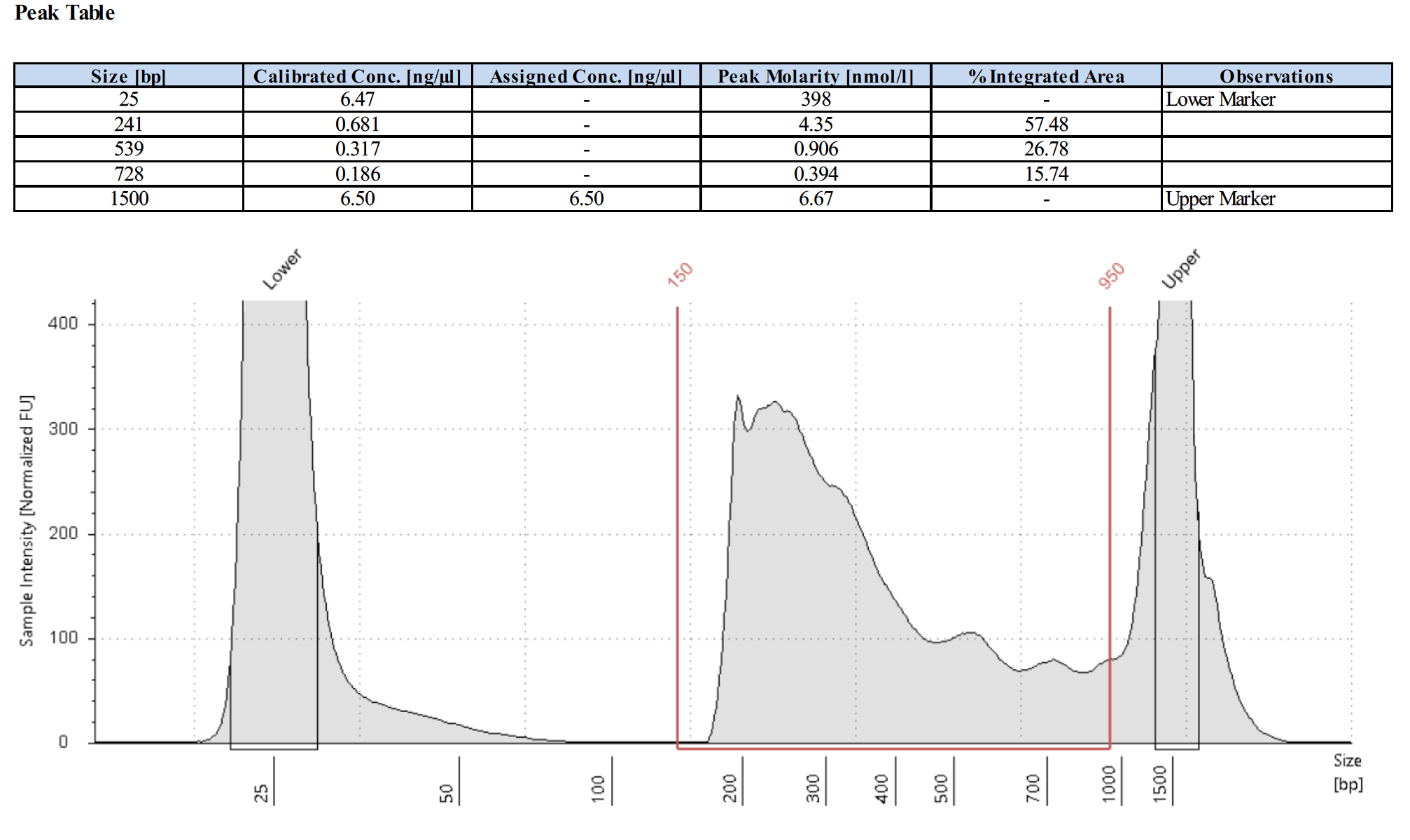
What do library traces look like for the negative control IgG?
Here are 2 examples of the negative control library traces.


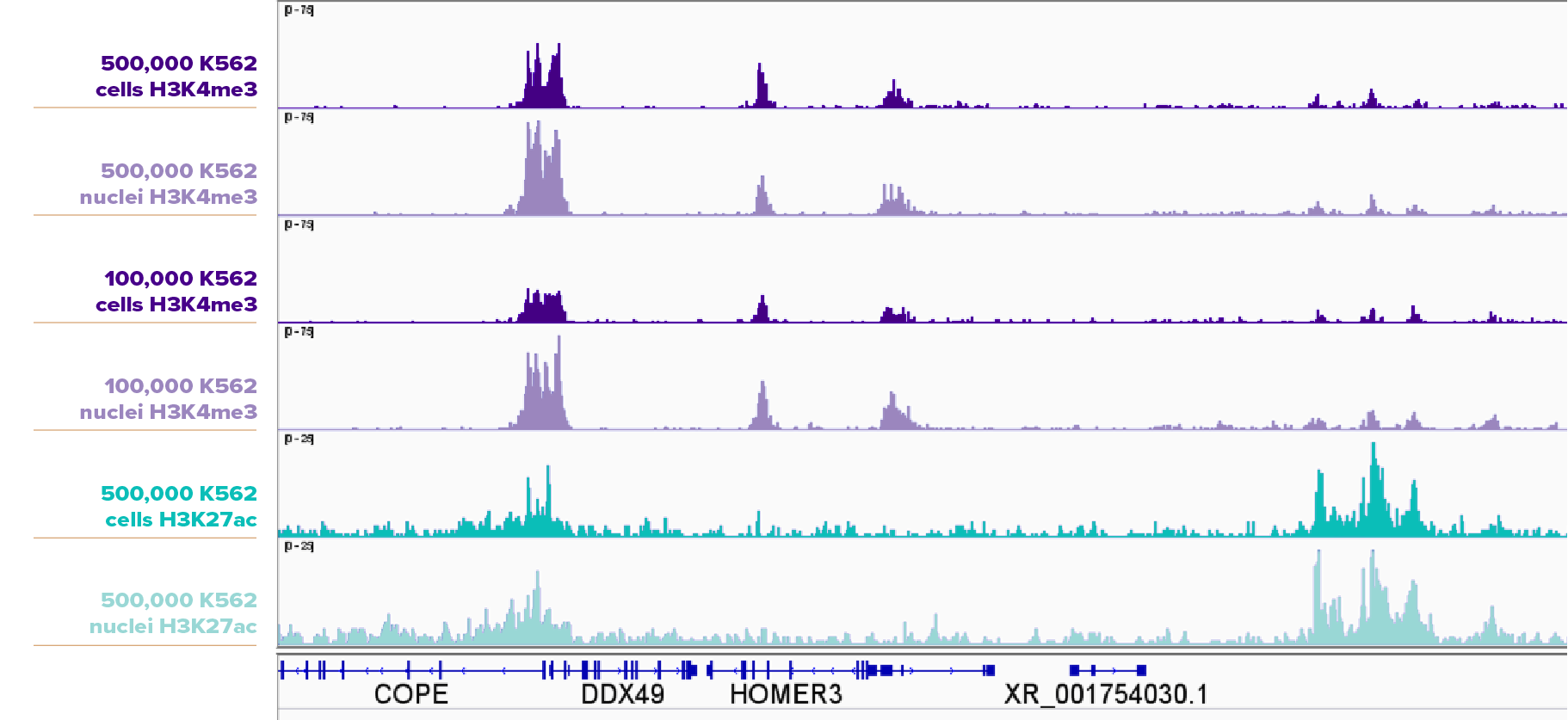
How do CUT&RUN results compare in cell samples versus nuclei samples?
We recommend nuclei, but in some cases the peak profiles were higher with cell samples for some transcription factor targets. Please see the figures below for details.
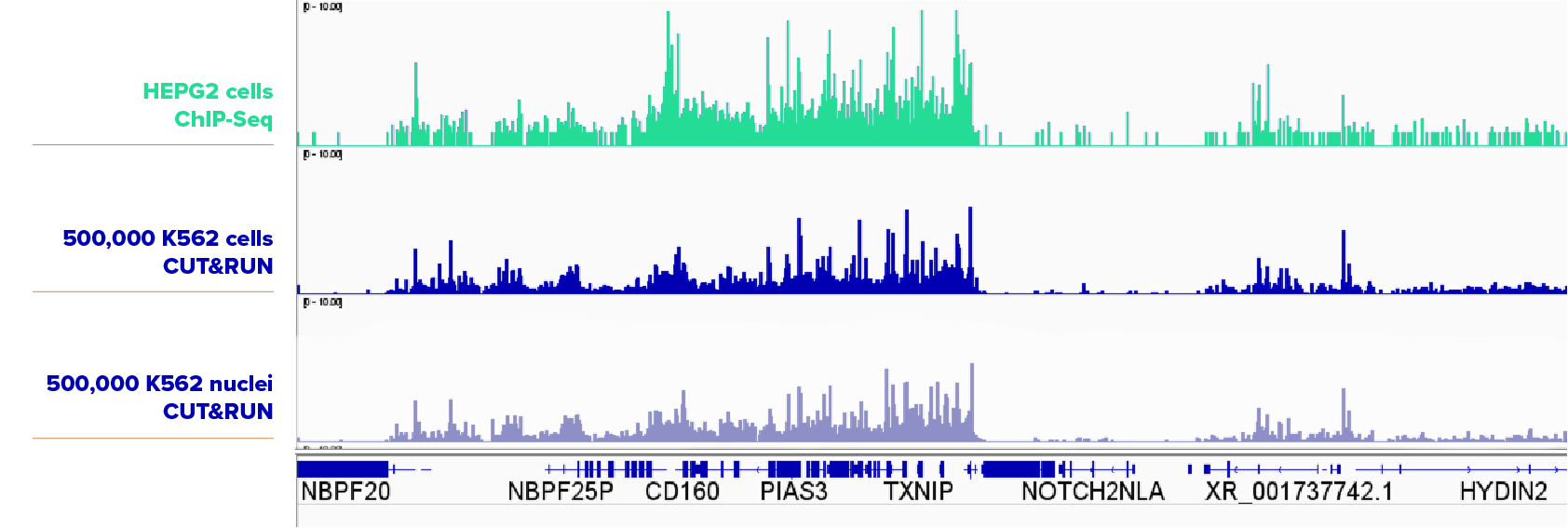
500,000 K562 cells and 500,000 K562 nuclei were assayed in CUT&RUN with the SUZ12 antibody (Cat No. 39357). The cell samples and nuclei samples yielded very similar results. The top track is ChIP-seq data from HEPG2 cells for comparison.
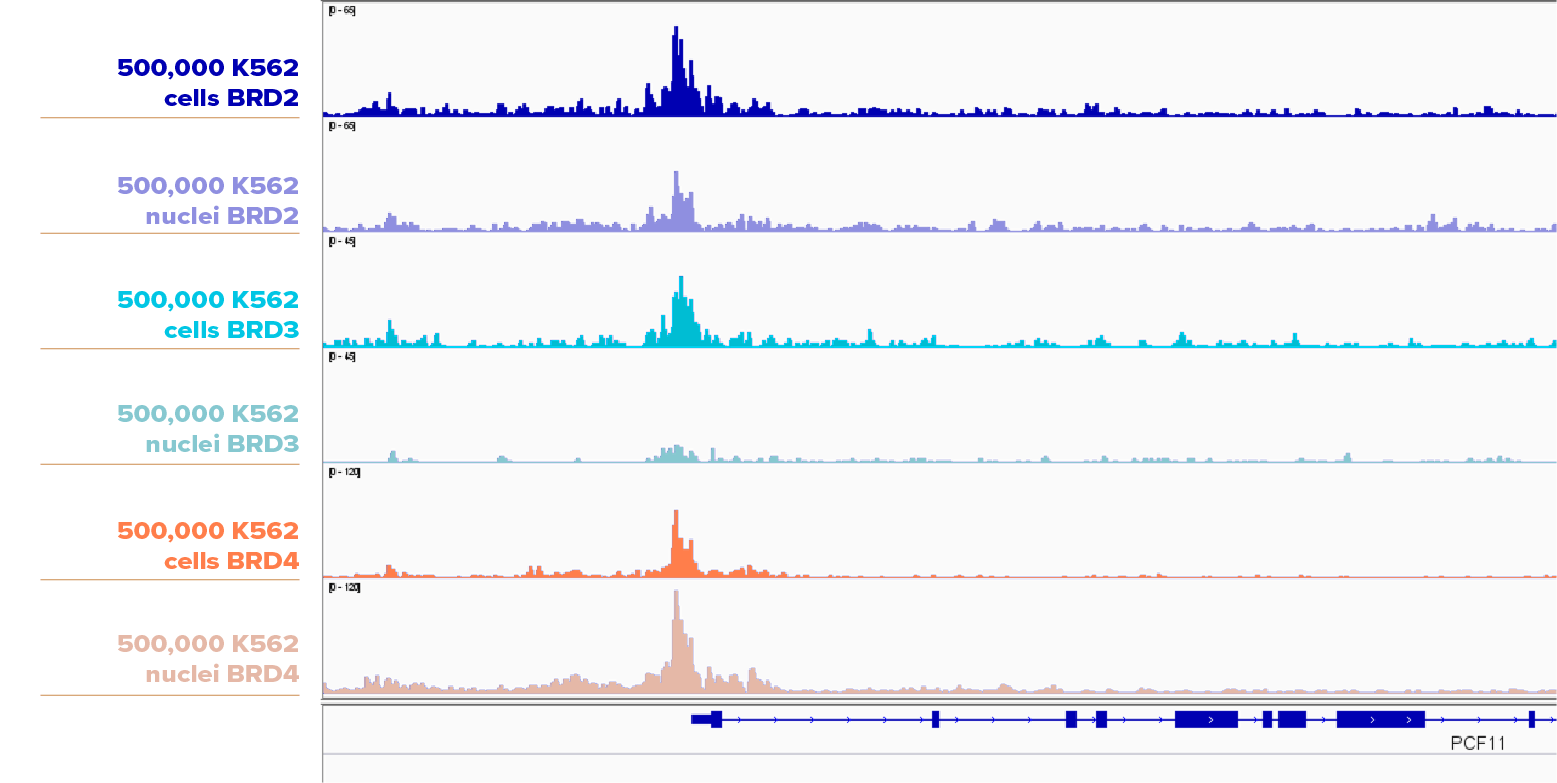
500,000 K562 cells and 500,000 K562 nuclei were assayed in CUT&RUN with BRD2 (Cat No. 61797), BRD3 (Cat No. 61489), or BRD4 (Cat No. 91301). The cell samples yielded slightly higher peaks than nuclei samples for BRD2 and BRD3, while the nuclei samples produced higher peaks than the cell samples for BRD4.
500,000 and 100,000 K562 cells and nuclei were assayed in CUT&RUN for H3K4me3 (Cat No. 91263), and 500,000 K562 cells and nuclei were assayed in CUT&RUN for H3K27ac (Cat No. 91193). The nuclei samples produced higher peaks than the cell samples.
Are ChIP-Seq or CUT&Tag-validated antibodies going to work with the CUT&RUN Assay Kit?
CUT&RUN, CUT&Tag, and ChIP-Seq have quite different workflows. If an antibody works for CUT&Tag or ChIP-Seq, that does not necessarily mean it will work in CUT&RUN. We advise using one our CUT&RUN-validated antibodies for CUT&RUN.
What does the library peak profile look like for YY1?
What does the library peak profile look like for JunD?
ChIC/CUT&RUN Assay Kit Documents