Recombinant Histones to study site- and degree-specific post-translational modifications. Our growing list of available recombinant histones include site- and degree-specific methylation, acetylation and phosphorylation. The combination of histone post-translational modifications form the basis of the 'histone code' that serves to regulate a variety of nuclear functions, such as interactions with chromatin-associated proteins, nucleosome remodeling, transcriptional regulation, replication and DNA repair. Each recombinant histone is prepared using one of two patented technologies: Expressed Protein Ligation (EPL) or Methylated Lysine Analog (MLA). We also offer a subset of our histone H3 proteins biotinylated for use in FRET assays.
With our MLA technology, methylated histones are generated via a chemical alkylation reaction to introduce a methyl-lysine analog at the desired lysine location, giving us precise control over the site and degree of methylation. Alternatively, the EPL technology can be used to generate methylated, acetylated and phosphorylated histones in which the histone globular domain is ligated to a peptide containing the N-terminal histone tail with site-specific modifications. This ligation reaction maintains the native histone bonds. For more information on EPL and MLA technologies, click on the EPL and MLA Technologies tab at the bottom of the page. All proteins are determined to be over 98% pure.
Active Motif has now added Recombinant Histone H3 biotinylated protein to it's portfolio of recombinant histones. The recombinant H3 protein is linked to biotin at the N-terminus via a carbon linker. The addition of the biotin enables capture of protein binding interactions utilizing the recombinant histone as a substrate. The biotinylated histone H3 protein is also an ideal substrate for homogenous FRET assays. Simply incubate the recombinant histone with the enzyme of interest and detect using streptavidin-coated donor beads and antibody-conjugated acceptor beads. Click on our link to learn more about the Recombinant Histone H3 biotinylated.
A complete list of recombinant histones is shown below. Click on the protein name to see complete information.
| Name | Expressed In | Format | Cat No. | Price | |
|---|---|---|---|---|---|
| Recombinant Histone H1 | E. coli | 100 µg | 81126 | $120 | Buy |
| 1 mg | 81826 | $545 | Buy | ||
| Recombinant Histone H1.2 | E. coli | 50 µg | 81252 | $175 | Buy |
| 1 mg | 81952 | $895 | Buy | ||
| Recombinant Histone H2A (Human) | E. coli | 100 µg | 31490 | $120 | Buy |
| 1 mg | 31890 | $545 | Buy | ||
| Recombinant Histone H2A/H2B dimer | E. coli | 100 µg | 81167 | $450 | Buy |
| 1 mg | 81867 | $3,000 | Buy | ||
| Recombinant Histone H2A.Z/H2B dimer | E. coli | 100 µg | 81168 | $465 | Buy |
| 1 mg | 81868 | $3,000 | Buy | ||
| Recombinant Histone H2B (Human) | E. coli | 100 µg | 31492 | $120 | Buy |
| 1 mg | 31892 | $520 | Buy | ||
| Recombinant Histone TH2B | E. coli | 100 µg | 31577 | $175 | Buy |
| 1 mg | 31977 | $895 | Buy | ||
| Recombinant Histone H2BFWT | E. coli | 100 µg | 31578 | $175 | Buy |
| Recombinant Histone H3T3ph (EPL) | E. coli | 25 µg | 31274 | $450 | Buy |
| Recombinant Histone H3K4ac (EPL) | E. coli | 25 µg | 31275 | $450 | Buy |
The ability to alter protein structure through the introduction of synthetic modifications is a powerful technique that can be utilized to dissect protein function and to facilitate the generation of research tools or therapeutic reagents. Chemical synthesis of peptides is a technically demanding process and also restricts the size of the generated protein. Therefore, a more robust technique is required to generate large modified proteins, such as histones.
Current methods used for constructing modified recombinant histones include solid phase peptide synthesis (SPPS), reductive alkylation and semisynthetic methods that utilize transferases to catalyze the transfer of modified groups to specified residues. However these methods have their disadvantages, including:
- Size limitation of the synthesized protein
- The availability of the transferase enzyme
- Variability in the degree of modifications catalyzed by the reaction
- Heterogeneity in the specificity of the enzyme
EPL and MLA Technologies
To overcome these limitations, Active Motif’s recombinant histones are engineered using one of two patented technologies: Expressed Protein Ligation (EPL) or Methylated Lysine Analog (MLA).
EPL is a semisynthetic method in which an expressed protein (rather than a synthesized peptide with limited fragment size) containing an unprotected C-terminal thioester is ligated to a peptide containing an N-terminal cysteine via a chemoselective reaction. The reaction, known as ‘native chemical ligation,’ preserves the native amine bond. In the case of recombinant histones, this involves the ligation of a histone tail containing the site-specific modification to the globular histone globular domain. Along with preserving the natural protein structure, this method allows for the engineering of large synthetic proteins and permits incorporation of a broad range of histone modifications found in nature.
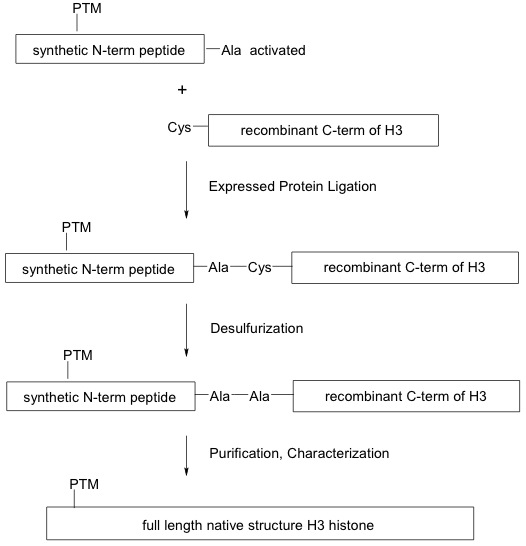
Click on image to enlarge size.
Figure 1: Generation of modified recombinant histones using the EPL method.
Expressed protein ligation (EPL) is used to synthesize full-length modified recombinant H3 histone proteins. A synthetic N-terminal peptide containing the post-translational modification (PTM) and an alanine (Ala)-conjugated thioester (activated) is ligated to a cysteine (Cys)-containing expressed C-terminal histone H3 globular domain through a series of reactions that preserve the native peptide bonds.
Using the alternative MLA technology, methylated histones are generated via a chemical alkylation reaction that substitutes a methylated analog of lysine, aminoethylcysteine, for the existing lysine at the desired residue. Aminoethylcysteine is structurally and chemically similar to lysine, though it contains an ethylamine substitution in place of the lysine ϒ-methylene. The lysine analogs can be chemically treated for engineering of modified histones to provide precise control over the site and degree of methylation. Studies demonstrate that modified histones engineered using the MLA technique show functional similarity to their natural counterparts.
Click on image to enlarge size.
Figure 2: Comparison of the chemical structures of lysine and methyl-lysine analogs.
The image depicts the similarities in the chemical structure of lysine and and the lysine analog, aminoethylcysteine. Aminoethylcysteine contains and ethylamine substitution (highlighted in gray) in place of the lysine ϒ-methylene (highlighted in gray). Alkylation of the ethylamine residue converts aminoethylcysteine into mono-, di- and tri-methyl lysine analogs (Me=methyl, in red).

Figure 3: Comparison of the performance of histone substrates in a histone demethylase assay reveal recombinant histone H3K4me2 protein (MLA) more closely mimics native histone substrates.
The positive control LSD1 enzyme from the Histone Demethylase Assay (Cat No. 53200) was used to assay for demethylase activity using either a histone H3K4me2 peptide or the included recombinant histone H3K4me2 (MLA) protein (Cat No. 31209). One µg of LSD1 was tested with either 70 µM H3K4me2 peptide or with 13 µM recombinant histone H3K4me2 protein generated using MLA technology. LSD1 was able to convert 73% of the recombinant histone H3K4me2 protein substrate into formaldehyde, yet it was only able to convert 14% of the H3K4me2 peptide into formaldehdye, even though there was 5-fold more peptide available than recombinant protein for the same amount of LSD1 enzyme. The higher rates of conversion achieved from the use of the recombinant histone H3K4me2 (MLA) protein more closely resemble in vivo conditions and demonstrate the high level of functional similarity between modified histones generated using MLA techniques and native histones.
References
- Lin, J.C. et al. (2007) Cancer Cell, 12: 432-444.
- Muir, T.W. (2003) Annu Rev Biochem, 72: 249-289
* The MLA technology is covered under U.S. Patent No. 8,278,112. EPL patent is pending.