This product has been discontinued and has been replaced by the MethylCollector MBD Capture Kit, cat# 55026
Active Motif's MethylCollector™ Ultra offers a fast magnetic assay capable of efficiently isolating methylated CpG islands from fragmented genomic DNA*. MethylCollector Ultra is based on the Methylated CpG Island Recovery Assay (MIRA) which uses a MBD2b/MBD3L1 protein complex for improved enrichment of CpG dinucleotides. Enriched DNA can be used in many downstream applications, such as end point or real time PCR analysis of the methylation status of particular loci in normal and diseased samples, bisulfite conversion followed by cloning and sequencing, or amplification and labeling for microarray analysis.
To learn more about how the kit works, please select the Description tab below. To see data and results, including comparisons to alternative methods, select the Data tab below.
Publication: A Versatile Assay for Detection of Aberrant DNA Methylation in Bladder Cancer. S Tommasi, A Besaratinia - Urothelial Carcinoma, 2018 - Springer
*Technology covered under U.S. Patent No. 7,425,415.
| Name | Format | Cat No. | Price | |
|---|---|---|---|---|
| MethylCollector™ Ultra | 30 rxns | 55005 | Discontinued | |
| MethylCollector™ Ultra Manual |
| MethylCollector & MethylCollector Ultra-Seq MOTIFvations June 13 Article |
| Products and Services to Study DNA Methylation |
| Epigenetics Products and Services |
MethylCollector Ultra Advantages
- High affinity binding provides greater enrichment than other MBD capture or antibody-based methods
- Fast magnetic bead based protocol is completed in less than 3 hours
- Specifically detects methylated CpGs from 1 ng - 1 µg of DNA fragmented by sonication or enzymatic digestion
- Includes positive control DNA and PCR primers to ensure success
- Eluted DNA is suitable for various downstream applications
Methylated CpG Island Recovery Assay (MIRA)
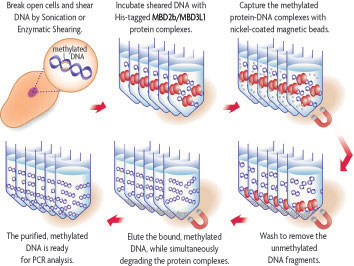
MethylCollector™ Ultra is based on the Methylated CpG Island Recovery Assay (MIRA) which utilize a His-tagged recombinant methyl-binding protein complex, comprised of MBD2b/MBD3L1, that specifically binds methylated CpGs of genomic DNA fragments that have been prepared by sonication or enzymatic digestion. Nickel-coated magnetic beads capture the protein-DNA complexes which are separated from the rest of the genomic DNA using the included magnet. Optimized buffers ensure that fragments with little or no methylation are removed. The methylated DNA is then eluted from the beads in the presence of Proteinase K. Following clean up, the eluted DNA is ready for use in PCR analysis or other downstream applications. See Flow Chart of MethylCollector™ Ultra process below.
The Methylated CpG Island Recovery Assay (MIRA) utilizes the high affinity of the MBD2b/MBD3L1 complex for methylated DNA1, to provide better enrichment than assays that use the methyl-binding protein MBD2 alone2. MethylCollector™ Ultra can efficiently enrich for methylated DNA from as few as 170 cells (~1 ng DNA).
Flow Chart of the MethylCollector™ Ultra process.
References
1. Rauch, T. and Pfeifer, G. (2005) Lab. Investigations, 85: 1172-1180
2. Jiang, C.L. et al. (2004) J. Bio. Chem., 279: 52456-52464
Preparing DNA Samples for Next-Gen Sequencing following MethylCollector Ultra Enrichment
Enriched methylated DNA obtained using MethylCollector Ultra can be used directly in Next-Gen Sequencing sample processing protocols.
If you intend to perform Next-Gen sequencing analysis following MethylCollector Ultra enrichment of methylated DNA, please refer to the MethylCollector Ultra manual and the Illumina ChIP-Seq DNA Sample Prep Kit for instructions on how to prepare your DNA samples for Next-Gen sequencing.
Click on image to enlarge size.
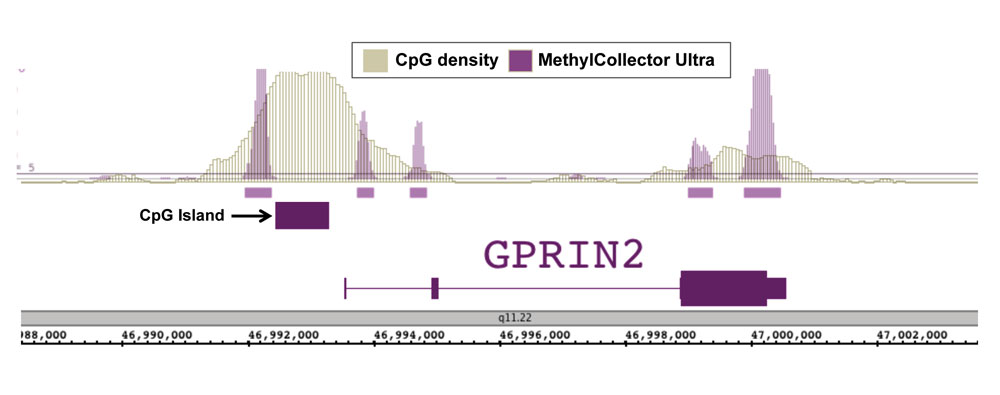
Figure 1: Next-Gen sequencing data generated using MethylCollector Ultra enriched DNA correlates well with CpG density.
DNA was enriched from 1 µg of human PBMC DNA using Active Motif’s MethylCollector Ultra Kit. Next-Gen sequencing was performed using the Illumina platform to generate 28 million sequence tags. Tags were mapped to generate a whole-genome DNA methylation profile. The image above shows that the enriched regions (purple peaks) correlate well with CpG density except at CpG islands, which should be largely unmethylated.
Click on image to enlarge size.
Figure 2: Next-Gen sequencing data generated using MethylCollector Ultra enriched DNA is more sensitive and robust than data from other MBD enrichment kits.
DNA was enriched from 1 µg of human PBMC DNA using Active Motif’s MethylCollector Ultra Kit and a competitor’s MBD-based enrichment kit. Next-Gen sequencing was performed using the Illumina platform to generate 28 million sequence tags. Tags were mapped to generate whole-genome DNA methylation profiles. The top purple panel is data produced from MethylCollector Ultra and the orange panel is data from the Competitor M kit. MethylCollector Ultra identified more methylated regions and the peaks had greater intensity than peaks identified from the competitor kit.
Click on image to enlarge size.
Figure 3: Next-Gen sequencing data generated using MethylCollector Ultra detects methylation at CpG shores.
DNA was enriched from 1 µg of human PBMC DNA using Active Motif’s MethylCollector Ultra Kit. Next-Gen sequencing was performed using the Illumina platform to generate 28 million sequence tags. Tags were mapped to generate a whole-genome DNA methylation profile. The image above shows one of many examples of DNA methylation being detected at CpG shores rather than in the CpG island itself. This data agrees with recent findings showing that a large portion of methylated sites occur in these CpG shore regions that are adjacent to CpG islands.
Preparing DNA Samples for Next-Gen Sequencing following MethylCollector Ultra Enrichment
Enriched methylated DNA obtained using MethylCollector Ultra can be used directly in Next-Gen Sequencing sample processing protocols.
If you intend to perform Next-Gen sequencing analysis following MethylCollector Ultra enrichment of methylated DNA, please refer to the MethylCollector Ultra manual and the Illumina ChIP-Seq DNA Sample Prep Kit for instructions on how to prepare your DNA samples for Next-Gen sequencing.
Click on image to enlarge size.
Figure 1: Next-Gen sequencing data generated using MethylCollector Ultra enriched DNA correlates well with CpG density.
DNA was enriched from 1 µg of human PBMC DNA using Active Motif’s MethylCollector Ultra Kit. Next-Gen sequencing was performed using the Illumina platform to generate 28 million sequence tags. Tags were mapped to generate a whole-genome DNA methylation profile. The image above shows that the enriched regions (purple peaks) correlate well with CpG density except at CpG islands, which should be largely unmethylated.
Click on image to enlarge size.
Figure 2: Next-Gen sequencing data generated using MethylCollector Ultra enriched DNA is more sensitive and robust than data from other MBD enrichment kits.
DNA was enriched from 1 µg of human PBMC DNA using Active Motif’s MethylCollector Ultra Kit and a competitor’s MBD-based enrichment kit. Next-Gen sequencing was performed using the Illumina platform to generate 28 million sequence tags. Tags were mapped to generate whole-genome DNA methylation profiles. The top purple panel is data produced from MethylCollector Ultra and the orange panel is data from the Competitor M kit. MethylCollector Ultra identified more methylated regions and the peaks had greater intensity than peaks identified from the competitor kit.
Click on image to enlarge size.
Figure 3: Next-Gen sequencing data generated using MethylCollector Ultra detects methylation at CpG shores.
DNA was enriched from 1 µg of human PBMC DNA using Active Motif’s MethylCollector Ultra Kit. Next-Gen sequencing was performed using the Illumina platform to generate 28 million sequence tags. Tags were mapped to generate a whole-genome DNA methylation profile. The image above shows one of many examples of DNA methylation being detected at CpG shores rather than in the CpG island itself. This data agrees with recent findings showing that a large portion of methylated sites occur in these CpG shore regions that are adjacent to CpG islands.
MethylCollector™ Ultra Contents & Storage
His-MBD2b/MBD3L1 protein complex, High Salt Binding Buffer, Low Salt Binding Buffer, Elution Buffer AM1, Protease Inhibitors, Proteinase K, Proteinase K Stop Solution, Magnetic Nickel Beads, Mse I digested Human, male genomic DNA, NBR2 PCR Primer Mix methylated control, Xist PCR Primer Mix methylated control, GAPDH PCR Primer Mix unmethylated control, Glycogen, PCR Buffer, Loading Dye, Bar Magnet, Glue dots and PCR tubes. Storage conditions vary from room temperature to -20°C, please see manual for specific details. All products guaranteed for 6 months from date of receipt.